Pharmacogenomics and Treatment of Neovascular AMD
How genotype can affect response to treatment.
Alan J. Franklin, MD, PhD • Sunil Gupta, MD • Magdalena Shuler, MD, PhD John Phillip Myers, MD • Brent Zanke, MD, PhD, FRCPC
Age-related macular degeneration is a genetically heterogeneous condition in which the mainstay of local anti-VEGF treatment also produces widely varied outcomes. Currently, there are only phenotypic predictors of response to anti-VEGF treatment,1,2 so we and many other groups have sought out genotyping as both a predictor for anti-VEGF treatment response and to identify novel potential targets for ocular antiangiogenic therapy.
Anti-VEGF treatment for neovascular AMD began with Macugen, a pegylated RNA aptamer used to block VEGF intraocularly.3,4 Subsequently, antibody-based technology, which utilized either a full-length monoclonal antibody, bevacizumab, or a truncated and affinity-purified monoclonal antibody, ranibizumab, was developed.5,6
These latter antibody technologies proved more effective than RNA aptamer blockage. However, for many years there were no class 1 studies to differentiate the efficacy and the safety of these two antibody-based molecules. Both the one- and two-year data from the CATT trial, as well as the one-year IVAN data, demonstrated that bevacizumab and ranibizumab affect visual acuity similarly when compared to each other in both monthly and as-needed maintenance schema.7-9
Eylea and novel antiangiogenic technologies that do not primarily target the VEGF protein will provide treating physicians with a wider variety of treatment options over the next several years.10 These advances can be significantly enhanced if there exists technology that can stratify and predict responses to different antiangiogenic treatments, as opposed to the current technology, which is based solely upon phenotypic and anatomic parameters.
GENETIC VARIATION IN TREATMENT RESPONSE
It has been long understood that there is a significant genetic component to AMD, based on family history studies. The advances in our ability to analyze the human genome have paralleled the advances in intraocular neovascular inhibitors. Single-nucleotide polymorphisms (SNPs) associated with AMD were first characterized in genes involved in the alternate complement pathway. Subsequently, disease-associated SNPs have been associated with the genes that are involved in tissue remodeling, as well as oxidative and cholesterol metabolism.11,12
By combining haplotype odds ratios of these multiple SNPs, smoking history, and drusen size, our ability to predict the risk of individuals with AMD to progress to advanced sight-threatening AMD is so advanced that it approximates the level of correlation between tobacco smoking and the development of lung cancer, with an odds ratio >17 (Figure 1).2,13-15
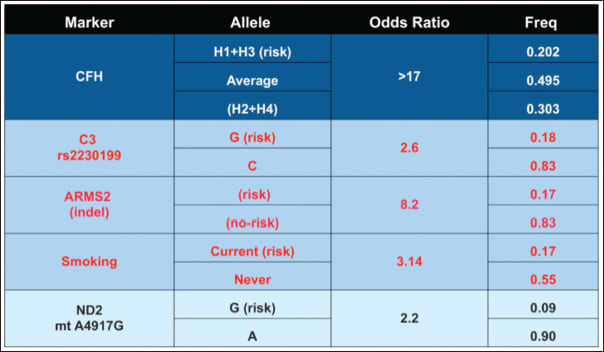
Figure 1. SNPs and haplotypes associated with AMD progression. Combining analysis of haplotypes and SNPs from four AMD-associated genes, combined with smoking history, provides an excellent predictability (OR = 0.83) reflex for the progression of AMD. The gene products utilized are involved in the complement pathway, CFH, C3, oxidative metabolism, ARMS2, and ND2. The two most significant genetic associations are in the CFH and ARMS2 genes.
To evaluate the potential association of various AMD SNPs with response to AMD treatment, it is important that the patient cohort evaluated responds to anti-VEGF in a similar fashion to the known, reported class 1 studies, such as the CATT and IVAN trials.7-9
Once the expected outcomes after an accepted antiangiogenic treatment paradigm for neovascular AMD can be verified, SNPs that are associated with AMD progression can be correlated with one- or two-year outcome data after anti-VEGF therapy to determine whether a rapid genetic analysis screen can predict response to treatment once a patient has developed neovascular AMD.
REPRODUCING THE CATT AND IVAN DATA
Approximately 600 patients who received anti-VEGF therapy for new-onset neovascular AMD were analyzed by retrospective chart review. Half the patients underwent anti-VEGF monotherapy with either bevacizumab or ranibizumab in a treat-and-extend or PRN protocol similar to that employed in the variable arm of the CATT trial.
The monotherapy arm was split almost equally between patients who received bevacizumab or ranibizumab. The eligibility criteria were similar to other AMD trials to include age older than 60 and new-onset macular extravasation secondary to AMD.
Lesions were comprised of less than 50% blood or fibrosis, while the exclusion criteria included optic atrophy or media opacity that precluded adequate retinal visualization, as well as previous adverse reactions to either triamcinolone acetonide or bevacizumab and patients who suffered a cerebrovascular accident within six months of initial presentation. These groups were then compared to each other by analyzing Snellen visual acuity, OCT, and treatment number data.
The retreatment criteria were also similar to the PRN arm of the CATT trial and were based on these analyses, so that evidence of persistent or recurrent extravasation on OCT was the primary determinant for retreatment. Patients in each group were similar in terms of age, presenting visual acuity, and sex.
The observed visual benefit was quite similar to that observed in the variable arm of the CATT trial, ie, 5.9 letters for bevacizumab and 6.8 letters for ranibizumab. Moreover, among these patients, there was no significant difference between the visual improvement in patients who received either anti-VEGF agent (P = .98), corroborating both the CATT and IVAN data.7-9
The required number of injections to achieve that visual gain observed after one year of anti-VEGF therapy was between six and seven, and there were no statistically significant differences in the required injection number between the anti-VEGF agents used, which parallels the required injection number of the first 12 months observed in the CATT trial, as well as the relative independence of injection number with respect to type of anti-VEGF agent used (Figure 2).
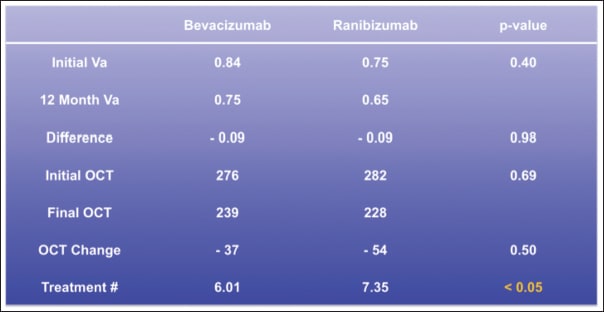
Figure 2. Comparison of visual acuity, OCT thickness, and treatment burden after 12 months of bevacizumab or ranibizumab administration. Both bevacizumab and ranibizumab achieved an average one-line gain in visual acuity after one year, and the P value was indicative of an almost identical visual result. The change in foveal thickness was also similar. Treatment burden was higher in patients who received ranibizumab.
Both agents effectively reduced central OCT thickness to a similar extent, with a trend toward ranibizumab being more effective at diminishing retinal thickness. Therefore, in a clinical practice, the variable arm of the CATT trial could be reproduced based on efficacy measures, such as visual acuity and reduction of central OCT thickness, as well as treatment burden.
GENETIC ASSOCIATION WITH ANTI-VEGF RESPONSE
Ninety patients among the group who responded typically to one year of anti-VEGF treatment underwent genetic analysis. DNA was harvested from peripheral blood lymphocytes by buccal swab, and 14 SNPs associated with the risk of AMD progression, which included the purported pathophysiological insertion/deletion polymorphism in the 3’ portion of the chromosome 10 ARMS2 gene, were studied.4
Based on the penetrance of these SNPs in the general population and history of tobacco smoking, an overall risk assessment has been created and verified to predict risk of progression to advanced dry or wet AMD. The overall risk assessment stratifies individuals with risk scores of 1 to 5. Categories 3-5 account for 20% of the population but represent 80% of cases of severe vision loss. Almost 50% of the general population is category 1.
Potential linkage to anti-VEGF response of this overall risk assessment score, which was previously determined to be predictive of AMD progression, was studied, in addition to linkage of the 14 individual AMD-associated SNPs. The average risk score in this patient cohort with neovascular AMD was 2.48, which affirms that patients who receive anti-VEGF treatment are generally at high genetic risk.
However, there was no correlation of overall risk score with response to anti-VEGF treatment in terms of visual acuity change, OCT thickness change, or treatment number. There were no independent associations of SNPs within the CFH, C3, or ND2 genes with response to anti-VEGF therapy based on these three parameters. However, at least one insertion/deletion polymorphic site, indel (NM_001099667.1:C.* 372_815del443ins54), within the ARMS2 gene identified by Hybe Probe Assay (Roche Applied Science), was found in 63 patients who had an average improvement in logMAR visual acuity of 0. 11, compared to a visual deterioration of 0.10 logMAR in those with the ancestral genotype (P = .0155, Wilcoxon rank sum test), which indicates both a clinically and statistically significant association (Figure 3).
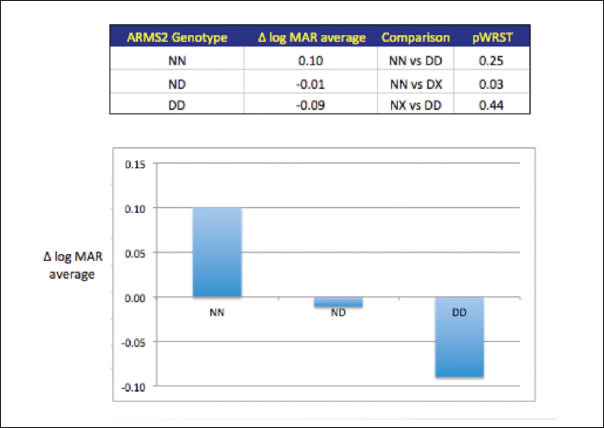
Figure 3. ARMS2 indel mutation is associated with visual protection. Analysis of the AMD associated SNPs and haplotypes in this figure show a significant correlation of the ARMS2 indel deletion with a better visual result following one year of anti-VEGF treatment with either bevacizumab or ranibizumab. Patients with the wild-type genotype typically lost 0.11 lines of vision, whereas the indel deletion imparted visual protection with 0.10 lines of average visual improvement. No significant associations were observed in treatment number or anatomic changes with the indel mutation or other AMD-related SNPs.
This indel polymorphism in the ARMS2 gene did not appear to be related to a significant difference in the treatment number or anatomic response based on OCT thickness. Patients with or without deletions did not differ by age (76.2 vs 76.4 years old, nonsignificant) or presenting visual acuity (0.77 vs 0.79, P = .84). The positive treatment response related to this mutation was similar in patients who were treated with either bevacizumab or ranibizumab.
ANTI-VEGF RESPONSE AND ARMS2 GENE STRUCTURE
Many groups have reported linkage of AMD markers that may predict response to anti-VEGF treatment and photodynamic therapy, which include genetic polymorphisms CC (rs1061170, Y402H) in the complement factor H gene (CFH)16,17 and the APOE gene.18 Variable response to AMD therapy was initially studied for PDT, in which visual acuity change was linked to the Y402H SNP in complement factor H.19-21
Subsequently, this SNP in the CFH gene was associated with variable response to intravitreal bevacizumab injected every six weeks, in which patients harboring the CC genotype lost visual acuity and patients with the TT or TC genotype gained vision.21 The protective effect of the TC and TT genotypes was supported by a larger study of almost 300 patients, in which those harboring the CC genotype had a higher risk of losing vision.16
There did not appear to be an association of this SNP with treatment burden, OCT response, or lesion size. Early visual response also appeared to be linked to an SNP in the APOE gene in a study of 200 patients.18
Analysis of a large cohort of patients in the CATT trial has not confirmed the linkage of any AMD-related SNPs to differential response of anti-VEGF therapy. These data describe the first report of a relationship between treatment response and polymorphisms within the LOC387715/ARMS2 gene.
The ARMS2 loci appear to have two gene products, LOC387715 and HTRA122,23. The LOC387715 gene is primate-specific and the gene product is purportedly related mitochondrial function, whereas the HTRA1 is a multifunctional serine protease.22,23
The LOC387715 gene is thought to downregulate expression of the HTRA1 gene. The indel mutation in the LOC387715 destabilizes the mRNA of the LOC387715 gene so that HTRA1 gene is upregulated, ultimately leading to AMD phenotype.24
More recent data suggest that the indel deletion in the LOC387715 gene is necessary, but not sufficient, to cause pathological genetic expression.17,25
Therefore, there appears to be complex control of the genes that comprise the ARMS2 loci, where changes in the genetic code can produce either loss or gain of function, ultimately resulting in variable phenotypes of protection or worsening of disease.
CONCLUSIONS
Two of the central issues in the treatment of neovascular AMD remain optimizing the results of anti-VEGF treatment with the lowest treatment burden. The next advances in treatment of neovascular AMD will likely be contingent upon the ability to predict the response of antiangiogenic treatment better and to develop neovascular inhibitors with novel mechanisms of action. Genetic analyses of patients undergoing anti-VEGF treatment have the potential to help to accomplish these two advances.
To confirm that the analysis was performed on wet AMD patients who responded typically to treatment, visual acuity, central OCT thickness, and treatment burden data were tabulated, and indeed, the results were strikingly similar to the one-year data from the PRN arm of the CATT and IVAN trials. In this cohort of patients, the monotherapy data with either ranibizumab or bevacizumab represent roughly 50% of the patients that enrolled in the variable arm of the CATT trial, and they were retrospective, rather than prospective, data.
Nevertheless, the close agreement of our data to those reported in the CATT study provides proof of concept that the CATT results can be reproduced in a standard private practice setting and that our genetic analysis was performed on patients whose response to anti-VEGF treatment paralleled those who underwent similar treatment in the CATT trial.
The reported relatively strong association of the indel deletion of the ARMS2 gene with vision protection following 12 months of anti-VEGF treatment, in a population that is representative of typical anti-VEGF response, provides a framework for the development of confirmatory prospective studies and for the possible design of novel ocular antiangiogenic strategies.
Pharmacogenomics of AMD is therefore a rapidly evolving field, and initial studies of patients with neovascular lesions have provided both interesting results and the rationale for larger studies to allow ultimately for better-individualized patient care and the design of new therapies. RP
REFERENCES
1. Age-Related Eye Disease Study Research Group. Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study Report Number 3. Ophthalmology. 2000;107:2224-2232.
2. Clemons TE, Milton RC, Klein R, Seddon JM, Ferris FL 3rd; Age-Related Eye Disease Study Research Group. Risk factors for the incidence of advanced age-related macular degeneration in the Age-Related Eye Disease Study (AREDS) AREDS report no. 19. Ophthalmology. 2005;112:533-539.
3. Chakravarthy U, Adamis AP, Cunningham ET Jr, et al. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology. 2006;113:1508 e1-25.
4. Gragoudas ES, Adamis AP, Cunningham ET Jr, Feinsod M, Guyer DR. Pegaptanib for neovascular age-related macular degeneration. N Engl J Med. 2004;351:2805-2816.
5. Brown DM, Kaiser PK, Michels M, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1432-1444.
6. Rosenfeld PJ, Brown DM, Heier JS, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419-1431.
7. Chakravarthy U, Harding SP, Rogers CA, et al. Ranibizumab versus bevacizumab to treat neovascular age-related macular degeneration: one-year findings from the IVAN randomized trial. Ophthalmology. 2012;119:1399-1411.
8. Martin DF, Maguire MG, Fine SL, et al. Ranibizumab and bevacizumab for treatment of neovascular age-related macular degeneration: two-year results. Ophthalmology. 2012;119:1388-1398.
9. CATT Research Group; Martin DF, Maguire MG, Ying GS, Grunwald JE, Fine SL, Jaffe GJ. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N Engl J Med. 2011;364:1897-1908.
10. Heier JS, Boyer D, Nguyen QD, et al. The 1-year results of CLEAR-IT 2, a phase 2 study of vascular endothelial growth factor trap-eye dosed as-needed after 12-week fixed dosing. Ophthalmology. 2011;118:1098-1106.
11. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385-389.
12. Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227-3236.
13. Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. European journal of human genetics. Eur J Hum Genet. 2009;17:100-104.
14. Reynolds R, Hartnett ME, Atkinson JP, Giclas PC, Rosner B, Seddon JM. Plasma complement components and activation fragments: associations with age-related macular degeneration genotypes and phenotypes. Invest Ophthalmol Vis Sci. 2009;50:5818-5827.
15. Seddon JM, Reynolds R, Yu Y, Daly MJ, Rosner B. Risk models for progression to advanced age-related macular degeneration using demographic, environmental, genetic, and ocular factors. Ophthalmology. 2011;118:2203-2211.
16. Kloeckener-Gruissem B, Barthelmes D, Labs S, et al. Genetic association with response to intravitreal ranibizumab in patients with neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52:4694-4702.
17. Yang X, Hu J, Zhang J, Guan H. Polymorphisms in CFH, HTRA1 and CX3CR1 confer risk to exudative age-related macular degeneration in Han Chinese. Br J Ophthalmol. 2010;94:1211-1214.
18. Wickremasinghe SS, Xie J, Lim J, et al. Variants in the APOE gene are associated with improved outcome after anti-VEGF treatment for neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52:4072-4079.
19. Brantley MA, Jr., Edelstein SL, King JM, et al. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to photodynamic therapy. Eye (Lond). 2009;23:626-631.
20. Feng X, Xiao J, Longville B, et al. Complement factor H Y402H and C-reactive protein polymorphism and photodynamic therapy response in age-related macular degeneration. Ophthalmology. 2009;116:1908-1912 e1.
21. Lee AY, Raya AK, Kymes SM, Shiels A, Brantley MA Jr. Pharmacogenetics of complement factor H (Y402H) and treatment of exudative age-related macular degeneration with ranibizumab. Br J Ophthalmol. 2009;93:610-613.
22. Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989-992.
23. Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992-993.
24. Fritsche LG, Loenhardt T, Janssen A, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892-896.
25. Yang Z, Tong Z, Chen Y, et al. Genetic and functional dissection of HTRA1 and LOC387715 in age-related macular degeneration. PLoS Genet. 2010;6:e1000836.
| Alan J. Franklin, MD, PhD, Sunil Gupta, MD, Magdalena Shuler, MD, PhD, and John Phillip Myers, MD, serve on the staff of the Retina Specialty Institute in Pensacola, FL. Brent Zanke, MD, PhD, FRCPC, is chairman and chief medical officer of Arctic Dx in Bonita Springs, FL. Drs. Franklin, Gupta, and Myers report financial interests as consultants to Arctic Dx, QLT, Novartis, Alcon, and RFE Pharma. Dr. Zanke reports significant financial interest in Arctic Dx. Dr. Shuler reports no financial interests in any products mentioned in this article. Dr. Franklin can be reached via e-mail at alfranklin@retinaspecialty.com. |








