PEER REVIEWED
Using Biomarkers for Retinal Disease Risk Assessment and Management
Identifying markers for AMD and diabetic retinopathy may spur new treatment options
Christine N. Kay, MD • Elliott H. Sohn, MD • Michael D. Abràmoff, MD, PhD
Biomarkers are proxies for biological and pathological processes that can be measured objectively. They are clinically useful when the biomarker precedes the disease process, thus serving as a risk factor, or when the biomarker acts as a surrogate for tissue or fluid that is not obtainable practically. Biomarkers can be used in the diagnosis and management of disease, such as using markers of blood pressure, cholesterol, and body mass index to determine the risk for cardiovascular disease.
Retinal biomarkers have become a target in research, because they can assist in elucidating mechanisms of disease, determine the risk of disease and disease progression, and serve as outcome parameters for clinical studies. This brief report will discuss genetic, biochemical, and structural biomarkers for two common retinal diseases: age-related macular degeneration and diabetic retinopathy.
AGE-RELATED MACULAR DEGENERATION
In AMD, genetic biomarkers are primarily used to estimate the risk of AMD and progression to choroidal neovascularization; biochemical markers are used to unravel the pathophysiology of AMD and CNV, and structural biomarkers are used both as risk estimators and for disease management.
Genetic
After blood is drawn from patients with AMD, DNA is extracted, and possible genetic associations are investigated using linkage disequilibrium studies and single nucleotide polymorphism (SNP) genotyping.
At present, the best available markers of AMD genetic risk are SNPs. SNPs in complement factor H (CFH) and PLEKHA1/ARMS2/HtrA1 capture a substantial fraction of AMD risk and permit the identification of individuals at high risk of developing AMD.1 Three groups simultaneously reported a CFH Y402H polymorphism, which appears to be a strong indicator of AMD predisposition. AMD risk for individuals bearing one variant allele increases by approximately 2.5-fold, while odds ratios (ORs) as high as 7.4 were reported for those patients who were homozygous for the risk allele.2-4 Several additional studies have confirmed a strong association between the Y402H SNP and AMD in the United States and have yielded population-attributable risk (PAR) estimates ranging from 43% to 68%.5-9
Further evidence implicating the complement system in AMD pathogenesis has come from two studies showing the existence of protective and risk haplotypes within the region containing BF and C2.10-11 A variant in the regulatory region of HtrA1 (rs11200638) (PLEKHA1/ARMS2/HtrA1) has also been associated with AMD.12-13 ORs for individuals heterozygous and homozygous for the risk alleles in the Caucasian population are 1.86 and 6.56, respectively, and the PAR for the HtrA1 SNP is 49.3% in the Caucasian population.12 Polymorphisms in these genes do not account for all patients with familial AMD; thus, current genotyping of patients with AMD is done more commonly for research than for clinical purposes at this point in time.
Biochemical
Various nongenetic tests are also being investigated to identify possible biomarkers of AMD. Most of these tests require a blood sample for serum analysis, with a particular interest in factors related to the inflammatory cascade; however, other noninvasive means of measuring nutrient levels are discussed below.
Inflammatory mediators. Proinflammatory cytokines are released from immune cells during an inflammatory response. Various immunological molecules and inflammatory mediators have been identified at the site of AMD lesions. Conflicting data exist regarding a possible link between C-reactive protein (CRP), a marker of non-specific inflammation, and AMD, as well as interleukin-6 (IL-6). Some researchers suggest that CRP may have a direct role in AMD development through its ability to induce complement activation,14 with another group describing a link between elevated CRP levels and AMD progression.15
However, other groups have demonstrated no significant association between CRP plasma levels and AMD.16 Similarly, Seddon's group described an association between IL-6 and AMD progression, yet Klein et al. found no such association.15-16
Finally, we know that VEGF plays a major role in angiogenesis and has become an important biological therapeutic target in the treatment of neovascular AMD. Some researchers have described significantly higher plasma VEGF levels in wet AMD patients compared with dry AMD patients, suggesting that high serum VEGF levels may play a role in predisposing individuals to neovascular AMD and may serve as an important biological biomarker in AMD.17
Nutrients. Given the ever-increasing studies on the in vivo effects of nutrients on AMD pathogenesis, serum levels of nutrients also show some promise in becoming important biomarkers. The randomized, placebo-controlled AREDS trial revealed that people at high risk for developing advanced stages of AMD lowered their risk by approximately 25% when treated for five years with a high-dose combination of vitamin C, vitamin E, b-carotene and zinc. The risk of vision loss caused by advanced AMD was reduced by approximately 19%.18
In the Eye Disease Case-Control Study of 421 cases and 615 control subjects, individuals with AMD with high serum levels of lutein plus zeaxanthin reduced the risk of neovascularization by a factor of two.19 However, there also exist a number of studies that have failed to reveal the association between serum levels of xanthophylls and AMD.20 Normal individuals have varying degrees of macular levels of xanthophylls, as measured by macular pigment optical density, yet the maculas of AMD patients reveal lower levels of macular pigment than age-matched controls.21 Furthermore, pathologic studies have shown that the macula in AMD patients has 62% of the lutein and zeaxanthin levels compared with the eyes of control subjects.22,52
It appears that using serum levels of xanthophylls as a biomarker for AMD may prove to be less effective than measuring macular levels using in vivo imaging. Because there are a number of noninvasive methods of measuring macular levels of lutein and zeaxanthin levels in vivo (flicker photometry, Raman spectroscopy, and fundus autofluorescence imaging), this method may allow adequate assessment of the risk of disease onset, progress of AMD, or the effects of treatment.
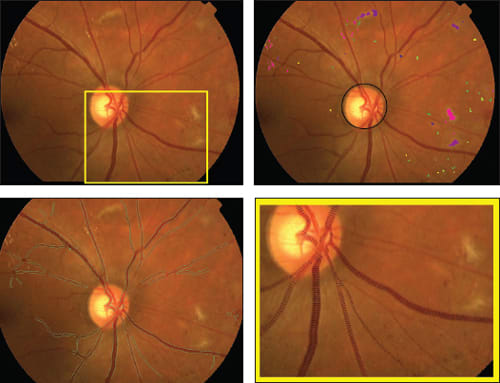
Figure 1. Fundus imaging biomarkers from a subject with diabetes and diabetic retinopathy. Input fundus color image (top left); lesion load metrics for hemorrhages (greenish), microaneurysms (yellowish), exudates (pinkish), retinal infarcts (purplish), the color shades indicate the confidence in the lesion type, and structural analysis of the disc location (top right); vessel analysis with green vessel boundaries (bottom left); detail of vessel analysis with local vessel widths visualized (bottom right).
Structural
Fundus and OCT images are obtained in patients with AMD, and the images are analyzed by experts or using computer programs to measure relevant biomarkers.23
Risk estimation. The type and quantity of drusen and the distribution of pigment in color fundus images are bio-markers for the risk of progression of AMD, as determined by reading centers.18 These lesions and their distributions can also be measured objectively and automatically using image analysis.24
Phenotype identification. Automated quantification of fundus phenotypes using image analysis allows differentiation between the fundus lesions in AMD and other macular diseases, such as Stargardt's disease. By quantifying the properties of the bright lesions (drusen and flecks) in their fundus images, these lesions can be distinguished with a computational algorithm. Quantification of fundus phenotypes may allow recognition of new phenotypes and correlation with new genotypes and may measure disease-specific biomarkers to improve the management of AMD patients.25
Disease management. With the advent of the anti-VEGF agents ranibizumab and bevacizumab, it has become clear that outer retinal and subretinal fluid is the main indicator of a need for anti-VEGF retreatment and that OCT can be used to predict disease activity and guide disease management.26-30
DIABETIC RETINOPATHY
In diabetic retinopathy, genetic biomarkers are being investigated to estimate the risk of developing DR and of progression to proliferative DR, biochemical markers are used to unravel the pathophysiology of DR and neovascularization, and structural biomarkers are used both as risk estimators and for disease management.
Genetic
The role of genetic factors in diabetic retinopathy is unclear but has long been a subject of interest. The search for genetic factors in a multifactorial, complex disease such as diabetes is characterized by two approaches: genome-wide scans for markers linked to disease and the study of candidate genes based on the putative function of the gene product.31
A large number of candidate genes have been examined in subjects with diabetes, but few groups have identified a strong association between a gene and the frequency or severity of retinopathy. Most published studies have been based on small patient samples and often are limited to a particular ethnic group.31 Immunoglobulin allotypes have been studied for possible associations with the progression of diabetic retinopathy, and an association between G2m(23) at the locus encoding IgG2 subclass heavy chains and predisposition to DR has been suggested.32
A relationship between DR and an insertion/deletion polymorphism in the angiotensin-converting enzyme gene in patients with type 2 diabetes has been reported.33 Other candidate genes of interest that have been studied include genes expressed in the aldose reductase pathway, glucose transporter genes, and endothelins and nitric oxide synthases (NOS).31 Studies on the NOS2A gene showed that a polymorphism of the NOS2A gene was protective (OR=0.21) against developing DR.34 A polymorphism of the aldose reductase gene has also been linked to the development of severe retinopathy in Caucasian subjects.35
Although many of these candidate genes have been linked with either lack of retinopathy or severe retinopathy, associations often cannot be replicated in multiple population groups. Genetic studies in a multifactorial disease can be challenging, but continued work is being done in this field, and candidate gene identification may become a valuable biomarker in the management of DR in the future.
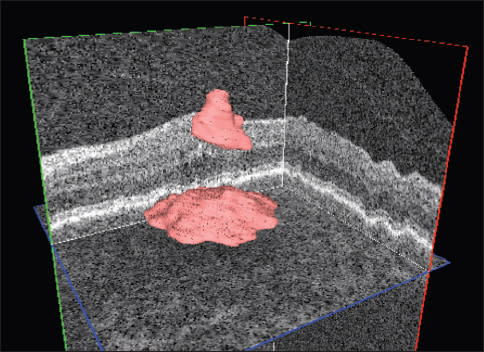
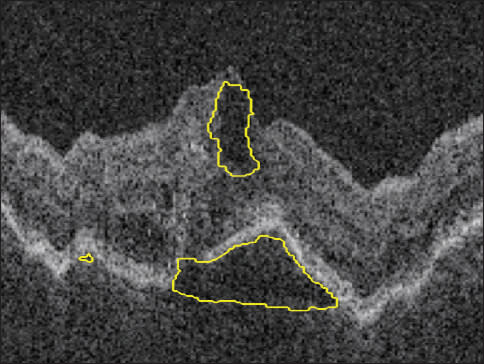
Figure 2. Automated quantification (above) and localization intraretinal and subretinal (below) fluid in a patient with AMD and choroidal neovascularization.
Biochemical
It is well known that glycemic control, diabetes duration and blood pressure are risk factors for microvascular complications in diabetes.36-39 More recently, other biomarkers have been investigated. In a cohort of the Diabetes Control and Complications Trial, the severity of retinopathy was associated with increasing triglyceride levels and inversely correlated with HDL levels.40
Some authors have suggested that serum apolipoprotein levels may be stronger biomarkers of DR than traditional lipid measures, such as total cholesterol, LDL or HDL cholesterol, or even the LDL-to-HDL ratio.41 The proteins apoAI (found in HDL) and apoB (found in LDL), and specifically the apoB-to-apoAI ratio, were significantly and independently associated with diabetic retinopathy development and severity.41
Other studies suggest a significant association between the plasma plasmin-α2-antiplasmin complex and severity of DR.42 Other studies have found a marked increase in protein oxidation in patients with diabetic microangiopathic complications (including retinopathy) through assessment of advanced oxidation protein products.43 Elevated levels of cytokines in ocular fluid may reflect disease activity but also reveal mechanisms of disease, such as in diabetic macular edema and AMD.44-45
Structural
Diabetic retinopathy has the longest history as a research subject in retinal image analysis. With the introduction of 3D OCT imaging and low-cost fundus imaging, a wealth of new information about the retinal morphology enabled its use for disease detection, understanding of pathophysiology, close monitoring and the guidance of retinal therapies.
Retinal neuropathy. Structural measurements, namely automated analyses of 3D OCT images of the retina, have shown that diabetes leads to a retinal neuropathy independent of vascular DR. In patients with diabetes who do not have DR or who have only minimal DR, the thickness of the ganglion cell layer in the macula is decreased in both type 1 and type 2 diabetes. These structural findings concur with animal models showing a loss of neural cells in diabetes. This retinal neuropathy is possibly related to diabetic neuropathies elsewhere in the body.46
Phenotype detection. Early detection of DR via population screening associated with timely treatment have been shown to prevent visual loss and blindness in patients with retinal complications of diabetes.47-48 Automated detection of image-based biomarkers, such as “red lesions,” ie, microaneurysms and hemorrhages, or “bright lesions,” ie, lipoprotein exudates and cotton-wool spots, allows early detection and risk assessment of patients at risk for DR, for example, in combination with telemedicine.49-50
Disease management. OCT is particularly useful in the management of DME, allowing detailed assessment of retinal thickness and morphologic evaluation of the neurosensory retinal layers.51 Currently, OCT imaging is widely used to determine the extent and volume of retinal thickening. We expect that detailed analyses of retinal layer morphology and texture from OCT will allow direct, image-based treatment to be guided by computer-supported or automated quantitative analysis of OCT.
CONCLUSION
Biomarkers in disease detection and management have become important tools in modern-day clinical medicine, and the field of retina is no exception. A combination of genetic markers, biochemical markers and structural analysis markers will likely become key components in the diagnosis and management of retinal disease. As more AMD and DR biomarkers are identified, a panel of them, in conjunction with select lifestyle factors, has the potential to be effective in identifying high-risk individuals, monitoring disease progression, and evaluating the efficacy of therapeutic intervention. Retina specialists should be aware of these biomarkers in order to optimally manage patients with common retinal diseases. RP
REFERENCES
1. Ross RJ, Verma V, Rosenberg KI, Chan CC, Tuo J. Genetic markers and biomarkers for age-related macular degeneration. Expert Rev Ophthalmol. 2007;2:443-457.
2. Edwards AO. Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. In: Science; 2005:308:421-424.
3. Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419-421.
4. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385-9.
5. Conley YP, Thalamuthu A, Jakobsdottir J, et al. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet. 2005;14:1991-2002.
6. Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A 2005;102:7227-7232.
7. Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77:389-407.
8. Magnusson KP, Duan S, Sigurdsson H, et al. CFH Y402H confers similar risk of soft drusen and both forms of advanced AMD. PLoS Med. 2006;3:e5.
9. Zareparsi S, Branham KE, Li M, et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet. 2005;77:149-`53.
10. Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055-1059.
11. Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458-462.
12. Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992-993.
13. Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989-992.
14. Seddon JM, George S, Rosner B. Progression of age-related macular degeneration: Prospective assessment of C-reactive protein, interleukin 6, and other cardiovascular biomarkers. Arch Ophthalmol. 2005;123:774-782.
15. Seddon JM, Gensler G, Milton RC, Klein ML, Rifai N. Association between C-reactive protein and age-related macular degeneration. JAMA. 2004;291: 704-710.
16. Klein R, Klein BE, Knudtson MD, Wong TY, Shankar A, Tsai MY. Systemic markers of inflammation, endothelial dysfunction, and age-related maculopathy. Am J Ophthalmol. 2005;140:35-44.
17. Tsai DC, Charng MJ, Lee FL, Hsu WM, Chen SJ. Different plasma levels of vascular endothelial growth factor and nitric oxide between patients with choroidal and retinal neovascularization. Ophthalmologica. 2006;220:246-251.
18. Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS Report 8. Arch Ophthalmol. 2001;119:1417-1436.
19. Eye Disease Case-Control Study Group. Antioxidant status and neovascular age-related macular degeneration. Arch Ophthalmol. 1993;111:104-109.
20. Mares-Perlman JA, Fisher AI, Klein R, et al. Lutein and zeaxanthin in the diet and serum and their relation to age-related maculopathy in the third national health and nutrition examination survey. Am J Epidemiol. 2001;153:424-432.
21. Bernstein PS, Zhao DY, Wintch SW, Ermakov IV, McClane RW, Gellermann W. Resonance Raman measurement of macular carotenoids in normal subjects and in age-related macular degeneration patients. Ophthalmology. 2002;109:1780-1787.
22. Bone RA, Landrum JT, Mayne ST, Gomez CM, Tibor SE, Twaroska EE. Macular pigment in donor eyes with and without AMD: a case-control study. Invest Ophthalmol Vis Sci. 2001;42:235-240.
23. Abramoff MD, Garvin M, Sonka M. Retinal imaging and image analysis. IEEE Rev in Biomed Engr. 2010;[in press].
24. Niemeijer M, van Ginneken B, Russell SR, Suttorp-Schulten MS, Abramoff MD. Automated detection and differentiation of drusen, exudates, and cotton-wool spots in digital color fundus photographs for diabetic retinopathy diagnosis. Invest Ophthalmol Vis Sci. 2007;48:2260-2267.
25. Quellec G, Russell SR, Scheetz TE, Stone EM, Abramoff MD. Computational quantification of complex fundus phenotypes in age-related macular degeneration and stargardt disease. Invest Ophthalmol Vis Sci. 2011;52:2976-2981.
26. Rosenfeld PJ, Moshfeghi AA, Puliafito CA. Optical coherence tomography findings after an intravitreal injection of bevicizumab (Avastin) for neovascular age-related macular degeneration. Ophthalmic Surg Lasers Imaging. 2005;36:331-335.
27. Heier JS, Antoszyk AN, Pavan PR, et al. Ranibizumab for treatment of neovascular age-related macular degeneration: a phase I/II multicenter, controlled, multi-dose study. Ophthalmology; 2006:113:633.e.1-4.
28. Preliminary report on effects of photocoagulation therapy. The Diabetic Retinopathy Study Research Group. Am J Ophthalmol. 1976;81:383-396.
29. Early Treatment Diabetic Retinopathy Study Research Group. Grading diabetic retinopathy from stereoscopic color fundus photographs—an extension of the modified Airlie House classification. ETDRS report number 10. Ophthalmology. 1991;98:786-806.
30. Harvey JN, Craney L, Nagendran S, Ng CS. Towards comprehensive population-based screening for diabetic retinopathy: operation of the North Wales diabetic retinopathy screening programme using a central patient register and various screening methods. J Med Screen. 2006;13:87-92.
31. Warpeha KM, Chakravarthy U. Molecular genetics of microvascular disease in diabetic retinopathy. Eye (Lond.). 2003;17:305-311.
32. Stewart LL, Field LL, Ross S, McArthur RG. Genetic risk factors in diabetic retinopathy. Diabetologia. 1993;36:1293-1298.
33. Nikzamir A, Rashidi A, Esteghamati A, Nakhjavani M, Golmohammadi T, Khalilzadeh O. The relationship between ACE gene insertion/deletion polymorphism and diabetic retinopathy in Iranian patients with type 2 diabetes. Ophthalmic Genet. 2010;31:108-113.
34. Warpeha KM, Xu W, Liu L, et al. Genotyping and functional analysis of a polymorphic (CCTTT)(n) repeat of NOS2A in diabetic retinopathy. FASEB J. 1999;13:1825-1832.
35. Demaine A, Cross D, Millward A. Polymorphisms of the aldose reductase gene and susceptibility to retinopathy in type 1 diabetes mellitus. Invest Ophthalmol Vis Sci. 2000;41:4064-4068.
36. Klein R, Klein BEK, Moss SE, Davis MD, DeMets DL. The Wisconsin Epidemiologic Study of Diabetic Retinopathy: II. Prevalence and risk of diabetic retinopathy when age at diagnosis is less than 30 tears. Arch Ophthalmol. 1984;102:520-526.
37. UK Prospective Diabetes Study Group: Tight blood pressure control and risk of macrovascular and microvascular complications in type 2 diabetes: UKPDS 38. BMJ. 1998;317:708-713.
38. Estacio RO, Jeffers BW, Gifford N, Schrier RW. Effect of blood pressure control on diabetic microvascular complications in patients with hypertension and type 2 diabetes. Diabetes Care. 2000;23(Suppl 2):B54-64.
39. Diabetes Control and Complications Trial Research Group. Progression of retinopathy with intensive versus conventional treatment in the Diabetes Control and Complications Trial. Ophthalmology. 1995;102:647-661.
40. Lyons TJ, Jenkins AJ, Zheng D, et al. Diabetic retinopathy and serum lipoprotein subclasses in the DCCT/EDIC cohort. Invest Ophthalmol Vis Sci. 2004;45:910-918.
41. Chait A, Montes VN. Apolipoproteins and diabetic retinopathy. Diabetes Care. 2011;34:529-531.
42. Nguyen TT, Alibrahim E, Islam FM, et al. Inflammatory, hemostatic, and other novel biomarkers for diabetic retinopathy: the multi-ethnic study of atherosclerosis. Diabetes Care. 2009;32:1704-1709.
43. Martín-Gallán P, Carrascosa A, Gussinyé M, Domínguez C. Biomarkers of diabetes-associated oxidative stress and antioxidant status in young diabetic patients with or without subclinical complications. Free Radical Biol Med. 2003;34:1563-1574.
44. Funk M, Schmidinger G, Maar N, et al. Angiogenic and inflammatory markers in the intraocular fluid of eyes with diabetic macular edema and influence of therapy with bevacizumab. Retina. 2010;30:1412-1419.
45. Campochiaro PA, Choy DF, Do DV, et al. Monitoring ocular drug therapy by analysis of aqueous samples. Ophthalmology. 2009;116:2158-2164.
46. van Dijk HW, Verbraak FD, Stehouwer M, et al. Association of visual function and ganglion cell layer thickness in patients with diabetes mellitus type 1 and no or minimal diabetic retinopathy. Vision Res. 2011;51:224-228.
47. Bresnick GH, Mukamel DB, Dickinson JC, Cole DR. A screening approach to the surveillance of patients with diabetes for the presence of vision-threatening retinopathy. Ophthalmology. 2000;107:19-24.
48. Kinyoun JL, Martin DC, Fujimoto WY, Leonetti DL. Ophthalmoscopy versus fundus photographs for detecting and grading diabetic retinopathy. Invest Ophthalmol Vis Sci. 1992;33:1888-1893.
49. Abramoff MD, Suttorp-Schulten MS. Web-based screening for diabetic retinopathy in a primary care population: the EyeCheck project. Telemed J EHealth. 2005;11:668-674.
50. Abramoff MD, Reinhardt JM, Russell SR, et al. Automated early detection of diabetic retinopathy. Ophthalmology. 2010;117:1147-1154.
51. Al-latayfeh MM, Sun JK, Aiello LP. Ocular coherence tomography and diabetic eye disease. Semin Ophthalmol. 2010;25:192-197.
52. Seddon JM, Ajani UA, Sperduto RD, et al. Dietary carotenoids, vitamins A, C, and E and advanced age-related macular degeneration. A multicenter study. JAMA. 1994;272:1413-1420.
| Christine N. Kay, MD, Elliott H. Sohn, MD, and Michael D. Abràmoff, MD, PhD, are all on the faculty of the Department of Ophthalmology and Visual Sciences and the Carver Family Center for Macular Degeneration at the University of Iowa in Iowa City. Dr. Abràmoff reports financial interest in patents and patent applications on retinal imaging and retinal biomarkers as owner of Idx, LLC. The other authors report no financial interest in any products mentioned in this article. Dr. Abràmoff can be reached via e-mail at michaelabramoff@uiowa.edu. |








